Exploratory Data Analysis: Kernel Density Estimation – Conceptual Foundations
June 9, 2013 34 Comments
For the sake of brevity, this post has been created from the first half of a previous long post on kernel density estimation. This first half focuses on the conceptual foundations of kernel density estimation. The second half will focus on constructing kernel density plots and rug plots in R.
Introduction
Recently, I began a series on exploratory data analysis; so far, I have written about computing descriptive statistics and creating box plots in R for a univariate data set with missing values. Today, I will continue this series by introducing the underlying concepts of kernel density estimation, a useful non-parametric technique for visualizing the underlying distribution of a continuous variable. In the follow-up post, I will show how to construct kernel density estimates and plot them in R. I will also introduce rug plots and show how they can complement kernel density plots.
But first – read the rest of this post to learn the conceptual foundations of kernel density estimation.
What is a Kernel?
Before defining kernel density estimation, let’s define a kernel. (To my surprise and disappointment, many textbooks that talk about kernel density estimation or use kernels do not define this term.)
A kernel is a special type of probability density function (PDF) with the added property that it must be even. Thus, a kernel is a function with the following properties
- non-negative
- real-valued
- even
- its definite integral over its support set must equal to 1
Some common PDFs are kernels; they include the Uniform(-1,1) and standard normal distributions. Wikipedia has a list of common kernels.
What is Kernel Density Estimation?
Kernel density estimation is a non-parametric method of estimating the probability density function (PDF) of a continuous random variable. It is non-parametric because it does not assume any underlying distribution for the variable. Essentially, at every datum, a kernel function is created with the datum at its centre – this ensures that the kernel is symmetric about the datum. The PDF is then estimated by adding all of these kernel functions and dividing by the number of data to ensure that it satisfies the 2 properties of a PDF:
- Every possible value of the PDF (i.e. the function,
), is non-negative.
- The definite integral of the PDF over its support set equals to 1.
Intuitively, a kernel density estimate is a sum of “bumps”. A “bump” is assigned to every datum, and the size of the “bump” represents the probability assigned at the neighbourhood of values around that datum; thus, if the data set contains
- 2 data at x = 1.5
- 1 datum at x = 0.5
then the “bump” at x = 1.5 is twice as big as the “bump” at x = 0.5.
Each “bump” is centred at the datum, and it spreads out symmetrically to cover the datum’s neighbouring values. Each kernel has a bandwidth, and it determines the width of the “bump” (the width of the neighbourhood of values to which probability is assigned). A bigger bandwidth results in a shorter and wider “bump” that spreads out farther from the centre and assigns more probability to the neighbouring values.
Constructing a Kernel Density Estimate: Step by Step
1) Choose a kernel; the common ones are normal (Gaussian), uniform (rectangular), and triangular.
2) At each datum, , build the scaled kernel function
,
where is your chosen kernel function. The parameter
is called the bandwidth, the window width, or the smoothing parameter.
3) Add all of the individual scaled kernel functions and divide by ; this places a probability of
to each
. It also ensures that the kernel density estimate integrates to 1 over its support set.
The density() function in R computes the values of the kernel density estimate. Applying the plot() function to an object created by density() will plot the estimate. Applying the summary() function to the object will reveal useful statistics about the estimate.
Choosing the Bandwidth
It turns out that the choosing the bandwidth is the most difficult step in creating a good kernel density estimate that captures the underlying distribution of the variable (Trosset, Page 166). Choosing the bandwidth is a complicated topic that is better addressed in a more advanced book or paper, but here are some useful guidelines:
- A small
results in a small standard deviation, and the kernel places most of the probability on the datum. Use this when the sample size is large and the data are tightly packed.
- A large
results in a large standard deviation, and the kernel spreads more of the probability from the datum to its neighbouring values. Use this when the sample size is small and the data are sparse.
There is a tremendous amount of literature on how best to choose the bandwidth, and I will not go into detail about that in this post. There are 2 methods that are common in R: nrd0 and SJ, which refers to the method by Sheather & Jones. These are string arguments for the option ‘bw’ in the density() function. The default in density() is bw = ‘nrd0’.
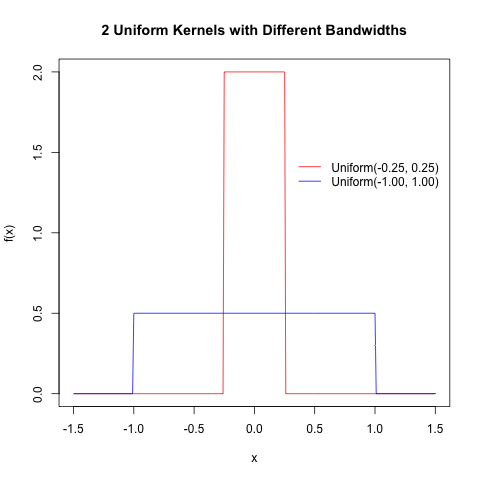
Here is some R code to generate the following plots of 2 uniform kernel functions; note the use of the dunif() function.
##### Plot 2 Uniform Kernel Functions with Different Bandwidths
##### By Eric Cai - The Chemical Statistician
# define support set of X
x = seq(-1.5, 1.5, by = 0.01)
# obtain uniform kernel function values
uniform1 = dunif(x, min = -0.25, max = 0.25)
uniform2 = dunif(x, min = -1.00, max = 1.00)
# optional printing of PNG image to chosen directory
png('INSERT YOUR DIRECTORY PATH HERE/uniform kernels.png')
plot(x, uniform1, type = 'l', ylab = 'f(x)', xlab = 'x', main = '2 Uniform Kernels with Different Bandwidths', col = 'red')
# add plot of second kernel function
lines(x, uniform2, col = 'blue')
# add legend; must specify 'lty' option, because these are line plots
legend(0.28, 1.5, c('Uniform(-0.25, 0.25)', 'Uniform(-1.00, 1.00)'), lty = c(1,1), col = c('red', 'blue'), box.lwd = 0)
dev.off()
References
Trosset, Michael W. An introduction to statistical inference and its applications with R. Chapman and Hall/CRC, 2011.
Everitt, Brian S., and Torsten Hothorn. A handbook of statistical analyses using R. Chapman and Hall/CRC, 2006.
Reblogged this on RANDOM THOUGHTS.
How can I make a density plot with discrete variables, for example, counts? Usually they have a poisson distribution, but the R density function density() does not have a kernel like this.
Hi Dominic,
Sorry for the late reply – I’ve been swamped at work and on hiatus from blogging.
By definition, a discrete random variable does not have a continuous distribution, so fitting a kernel density estimator for a discrete random variable does not make sense. It is supposed to be discrete; if your random variable has a Poisson distribution, then a histogram would be best at visualizing it.
Great suff Eric! Learned a bunch from this post! Was good to run into you today. I’ll make sure follow you 🙂
It was nice to see you, too, Fong! Thanks for visiting my blog!
Reblogged this on nishant@analyst.
Hi Eric, May I know why the mean and median for the kernel densitiy estimates on the data are the same? I used the function density () in R, and get the summary..it shows the same number of median and mean. May I know why please. However after reading your post , really clear my mind about Kernel density estimation…thanks a lot!
Hi sheeda – I think that you are referring to the part of the output for density() that has the heading “x”. Note that “x” is NOT the data set. As R’s documentation page for density() states, it is “the n coordinates of the points where the density is estimated”. These points are uniformly distributed along the range of “x”, so the mean and median of “x” are equal. If you want to find the mean and median of your data set, use the mean() and median() functions, respectively.
Hi Eric,
Thanks for replying…Now I understand that part. However, if x is the coordinates of the points where the density is estimated, how to get values of the kernel density estimate for an instance from the density () function? I am doing classification using Bayes theorem (certainly it is not in the topic that u discussed), by using Kernel estimation. Then, I need to find the posterior probability . To get the posterior probability I need the conditional probability that is estimated using Kernel density. Please guide me. God bless you..
Thanks Eric.
sheeda – I suggest that you read the documentation page for density() and learn how to extract the KDE from the output. Here is an example.
kde.output = density(rnorm(100))
kde.values = kde.output$y
Thanks Eric..its so simple, you saved my life!
How can we give kernel density estimation as a input to sampling methods in R? For example I want to sample from bi-variate kernel density distribution using ARMS sampling technique available in R in “HI” package?
Sorry Shruti – I don’t understand your question, and I’m not familiar with ARMS sampling and the “HI” package.
If you look at my other blog post on kernel density estimation, you can see that the KDE (specifically, the y-values and the x-values) can be stored as variables. Perhaps you can use those as inputs for your next task.
Hope that helps!
Hi,i have a problem:i use the function density in R for kernel density estimation,but it has values greater than 1.is it possible?where is the problem?
Hi Lorenzo,
When you say “it has values greater than 1”, what do you mean by “it”?
If you mean the density estimate, then there is no problem – it is entirely possible for a kernel estimate to exceed 1. The area under the curve needs to be equal to 1, but the value of a probability density function can definitely exceed 1.
Pingback: Where's Waldo? Algorithm Helps You Find Him : Discovery News
Pingback: Tim Brock
I have never had the chance to read a better definition of the kernel estimate! Simply amazing!
Thanks, Daniel – much appreciated!
Hi Eric, is a non parametric method better than a parametric one when calculating confidence intervals of time-series data?
Hi Tamas,
I’m sorry – I’m not an expert in time series analysis, so I can’t answer your question. However, I can give you some guidelines on how to frame your question.
First, we need to define “better” – in other words, we need to define the quality of a confidence interval. Statisticians usually judge an estimator by the bias and the variance. (Another term for “confidence interval” is “interval estimator”, so it is an estimator with a bias and a variance.)
Thus, if you want to compare a parametric CI with a non-parametric CI in any context, you need to assess those 2 things. My guess is that the bias is higher and the variance is lower for a parametric CI, because it makes more assumptions about the data.
It’s a small detail, but I appreciate that you made the “INSERT YOUR DIRECTORY PATH HERE/” part red. I wish I could format all my code that way.
You’re welcome, Gene! This red formatting can only be done in the text editor on my blog; there is no way to do this within R or RStudio, as far as I know.
#Espero sea de su agrado
##############################################
##Densidad de Kernel utilizando arreglos en r#
##############################################
##1.- Puntos observados
set.seed(10)
X=cbind(x=runif(n=100,min=0,max=50),y=runif(n=100,min=0,max=50))
##2.- Elejimos el rango para establecer los puntos donde se hará la estimación
x.p=seq(0,50,.2)
y.p=seq(0,50,.2)
##3.- Definimos el ancho de la ventana y la cantidad de datos
h=2.8
n=length(X[,1])
##Establecemos la funcion de kernel Epanechnikov
##Source: Nonparametric and Semiparametric Models (Wolfgang Härdle,Marlene Müller,Stefan Sperlich,Axel Werwatz)
k.e=function(u){0.75*(1 – u^2)*I(abs(u)<=1)}
########################
##Caso univariado
########################
m.k=outer(x.p,X[,1],function(x,y,h)
{u=(x-y)/h
k.e(u)},h)
d.k=apply(m.k,1,FUN=function(x){(1/(n*h))*sum(x)})
par(mfrow=c(1,2))
plot(x.p,d.k,type="l")
plot(density(X[,1],kernel="epanechnikov",adjust=.1,bw=h*4,n=101,
from=0,to=50))
#graphics.off()
###############################
##Caso bivariado###############
###############################
m.k.x=outer(x.p,X[,1],function(x,y,h)
{u=(x-y)/h
k.e(u)},h)
m.k.y=outer(y.p,X[,2],function(x,y,h)
{u=(x-y)/h
k.e(u)},h)
Z=(1/(n*h^2))*m.k.x%*%t(m.k.y)
##Grafico de la densidad bivariada
image(x=x.p,y=y.p,z=Z)
points(X)
Hi Eric,
Great post, it was a useful introduction to density estimation. I have a few questions though:
1. This may seem silly, but how do you tell whether a random variable is discrete or continuous? For instance, if I have an RV that only takes on values that are natural numbers, does that automatically make it discrete? Height for instance can be captured in inches, centimetres, or millimetres. What is an example of a continuous random variable, and does the categorisation depend on the resolution at which the RV is captured?
2. What are the criteria for picking a particular kernel function? R’s density uses the Gaussian kernel by default, but it’s not clear to me when to use a Gaussian vs Triangular vs Uniform. Any tips would greatly appreciated!
1. A discrete random variable has a *countable* number of possible values. Yes, a random variable that can take on only natural numbers is discrete.
A continuous random variable has an infinite number of possible values. Yes, height is a continuous random variable.
I don’t understand what mean by “the resolution at which the RV is captured”; please clarify.
2) I like using the normal (Gaussian) kernel function, because it is symmetrical, continuous, and has nice mathematical properties. Using a kernel function like the uniform or the triangular function could result in discontinuities in the estimated density.
Thanks for the reply, Eric. What I meant by resolution is this: let’s say I have 2 (hypothetical) tape measures, one that measures only in metres, and the other that measures in millimetres. Clearly, the latter will give me a more ‘precise’ estimate of my height than the former because of how fine-grained its divisions are.
Does this make height as measured by the first tape measure (in metres) a discrete variable (since it can only take on a few values), and the other height (measured in millimetres) a continuous random variable?
My take is that since we’re measuring the same quantity, it should be continuous, but I’m getting tripped up by the fact that discrete random variables only take on a few values.
Am I worrying too much about the definition? Thanks for your help!
Length is a continuous variable regardless of the unit that you use to measure it. Technically speaking, your height could be 1.95 metres or 195 centimetres, so measuring in metres does not *necessarily* result in less precision.
I understand what you mean, though – there could be round-off errors because of the imprecision (1.90 metres is rounded to 2 metres), but that doesn’t change the fact that length is still a continuous variable.
Reblogged this on Uma certa pressa …..
Readers of this post may be interested in the non-parametric density function density.fd proposed by Jim Ramsey in his fda R package. In this case, a roughness penalty controls the smoothness of the density function estimate.
Se for example: https://www.rdocumentation.org/packages/fda/versions/2.4.7/topics/density.fd
Thanks for sharing, Pascal!
Thanks for defining what is a ” kernel ” . May god bless you.
You’re welcome! It frustrated me a lot when I first encountered this term but found no definition. I’m glad that I could help you.
Pingback: Correlations – HuntDataScience