Exploratory Data Analysis: Combining Box Plots and Kernel Density Plots into Violin Plots for Ozone Pollution Data
June 16, 2013 9 Comments
Introduction
Recently, I began a series on exploratory data analysis (EDA), and I have written about descriptive statistics, box plots, and kernel density plots so far. As previously mentioned in my post on box plots, there is a way to combine box plots and kernel density plots. This combination results in violin plots, and I will show how to create them in R today.
Continuing from my previous posts on EDA, I will use 2 univariate data sets. One is the “ozone” data vector that is part of the “airquality” data set that is built into R; this data set contains data on New York’s air pollution. The other is a simulated data set of ozone pollution in a fictitious city called “Ozonopolis”. It is important to remember that the ozone data from New York has missing values, and this has created complications that needed to be addressed in previous posts; missing values need to be addressed for violin plots, too, and in a different way than before.
The vioplot() command in the “vioplot” package creates violin plots; the plotting options in this function are different and less versatile than other plotting functions that I have used in R. Thus, I needed to be more creative with the plot(), title(), and axis() functions to create the plots that I want. Read the details carefully to understand and benefit fully from the code.
Read further to learn how to create these violin plots that combine box plots with kernel density plots! Be careful – the syntax is more complicated than usual!
Violin Plots in R
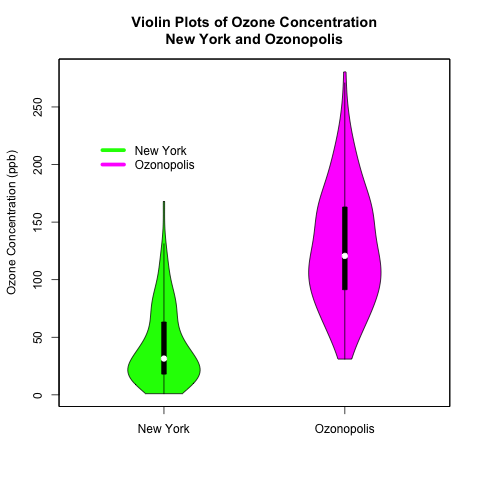
A violin plot is a box plot with a kernel density plot rotated and surrounding it on each side. In R, the vioplot() command, which is what I will use to create violin plots in this post, does not show the outliers like a regular box plot, and it does not allow the user to choose the type of kernel or the bandwidth. (Compare the violin plots at the end of this post with the box plots and kernel density plots in my previous posts to see the differences.)
To construct violin plots in R, install the “sm” and “vioplot” packages. (Depending on your R interface, this can be done via a menu, but it can always be done with code.) The “vioplot” package needs the “sm” package to work. The vioplot() command, which creates the violin plots, is part of the “vioplot” package.
Unfortunately, the vioplot() command does not have options to set a title or axis labels. Thus, I used a combination of the plot(), title(), and axis() functions to label my violin plots.
First, let’s get the data.
##### Violin Plots ##### By Eric Cai - The Chemical Statistician # extract "Ozone" data vector for New York ozone = airquality$Ozone # calculate the number of non-missing values in "ozone" n = sum(!is.na(ozone)) # calculate mean, variance and standard deviation of "ozone" by excluding missing values mean.ozone = mean(ozone, na.rm = T) var.ozone = var(ozone, na.rm = T) sd.ozone = sd(ozone, na.rm = T) # simulate ozone pollution data for ozonopolis # set seed for you to replicate my random numbers for comparison set.seed(1) ozone2 = rgamma(n, shape = mean.ozone^2/var.ozone+3, scale = var.ozone/mean.ozone+3)
Now, install the “sm” and “vioplot” packages. Then call the respective libraries.
library(sm) library(vioplot)
The vioplot() function does not have the option of ignoring missing values. (i.e. Unlike many other plotting functions in R, it does not have the “na.rm = TRUE” option.) Thus, for the ozone data set for New York, I obtained the indices of the non-missing elements of the “ozone” vector, and then I used those indices to extract a new vector of just the non-missing elements.
# extract new vector of non-missing elements of the "ozone" data from New York ozone.ny.na.rm = ozone[!is.na(ozone)]
Finally, construct the violin plots. As usual, I sandwich the plotting commands with png() and dev.off() to export a PNG image to my local directory. Read my comments for each line of code very carefully to understand why I used them.
png('INSERT YOUR DIRECTORY PATH HERE/violin plots.png')
# create a blank plotting frame with 1 row and 1 column
# I chose 4 units for xlim to leave plenty of space between the 2 violin plots
# ylim is set to cover all possible values of both ozone.ny.na.rm and ozone2
# type = "n" means no plotting - keep it blank
# xlab and ylab are left intentionally blank
# xaxt = 'n' means no tick marks
plot(1, 1, xlim = c(0, 4), ylim = range(c(ozone.ny.na.rm, ozone2)), type = 'n', xlab = '', ylab = '', xaxt = 'n')
# construct violin plots
# use separate command for each data set to customize individual colours
# "at =" sets the point along the horizontal axis where the vioplot is made
# "add = T" means that this plot is adding to a previously existing plot
vioplot(ozone.ny.na.rm, at = 1, add = T, col = 'green')
vioplot(ozone2, at = 3, add = T, col = 'magenta')
# set horizontal axis
axis(1, at = c(1,3), labels = c('New York', 'Ozonopolis'))
# set vertical axis
# at = 150 sets height of label at y = 150
# pos = -0.45 ensures that the label is at x -0.45 and not overlapping the numbers along the tick labels
# tck = 0 ensures that the label does not get its own tick mark
# try not including the at, pos, and tck options and see what you get
axis(2, at = 150, pos = -0.45, tck = 0, labels = 'Ozone Concentration (ppb)')
# add title
title(main = 'Violin Plots of Ozone Concentration\nNew York and Ozonopolis')
# add legend
# the option bty = 'n' ensures that there is no border around the legend
# the option lty must be specified, because I'm drawing lines as the markers for each city in the legend
# the option lwd specifies the width of the lines in the legend
legend(0.4, 250, legend = c('New York', 'Ozonopolis'), bty = 'n', lty = 1, lwd = 7, col = c('green', 'magenta'))
dev.off()
Here is the resulting plot!
Reblogged this on RANDOM THOUGHTS.
Pingback: How do I become a data scientist? | lordtomriddle
Reblogged this on nishant@analyst.
Pingback: How do I become a data scientist? | reverentiallyirreverent
Pingback: How to Become a Data Scientist | What a Computational Biologist Do?
Pingback: Mo-Data. Data made EasyDo you need to quickly become a data scientist? Great guide for self starters #datascience #bigdata - Mo-Data. Data made Easy
Pingback: How do i become a Data Scientist – Primose Training
Pingback: Do you need to quickly become a data scientist? Great guide for self starters #datascience #bigdata | MoData
Pingback: How can I become a data scientist? | Since 1989