Update on July 15, 2013:
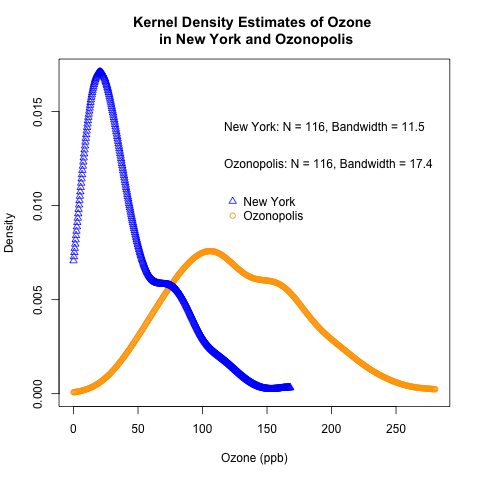
Thanks to Harlan Nelson for noting on AnalyticBridge that the ozone concentrations for both New York and Ozonopolis are non-negative quantities, so their kernel density plot should have non-negative support sets. This has been corrected in this post by
– defining new variables called max.ozone and max.ozone2
– using the options “from = 0” and “to = max.ozone” or “to = max.ozone2” in the density() function when defining density.ozone and density.ozone2 in the R code.
Update on February 2, 2014:
Harlan also noted in the above comment that any truncated kernel density estimator (KDE) from density() in R does not integrate to 1 over its support set. Thanks to Julian Richer Daily for suggesting on AnalyticBridge to scale any truncated kernel density estimator (KDE) from density() by its integral to get a KDE that integrates to 1 over its support set. I have used my own function for trapezoidal integration to do so, and this has been added below.
I thank everyone for your patience while I took the time to write a post about numerical integration before posting this correction. I was in the process of moving between jobs and cities when Harlan first brought this issue to my attention, and I had also been planning a major expansion of this blog since then. I am glad that I have finally started a series on numerical integration to provide the conceptual background for the correction of this error, and I hope that they are helpful. I recognize that this is a rather late correction, and I apologize for any confusion.
For the sake of brevity, this post has been created from the second half of a previous long post on kernel density estimation. This second half focuses on constructing kernel density plots and rug plots in R. The first half focused on the conceptual foundations of kernel density estimation.
Introduction
This post follows the recent introduction of the conceptual foundations of kernel density estimation. It uses the “Ozone” data from the built-in “airquality” data set in R and the previously simulated ozone data for the fictitious city of “Ozonopolis” to illustrate how to construct kernel density plots in R. It also introduces rug plots, shows how they can complement kernel density plots, and shows how to construct them in R.
This is another post in a recent series on exploratory data analysis, which has included posts on descriptive statistics, box plots, violin plots, the conceptual foundations of empirical cumulative distribution functions (CDFs), and how to plot empirical CDFs in R.

Read the rest of this post to learn how to create the above combination of a kernel density plot and a rug plot!
Read more of this post
and
, from the normal distribution
, where
is unknown. Then the statistic
.
does not depend on
, so
is an ancillary statistic for
.
is unknown, then
is not ancillary for
.

Recent Comments