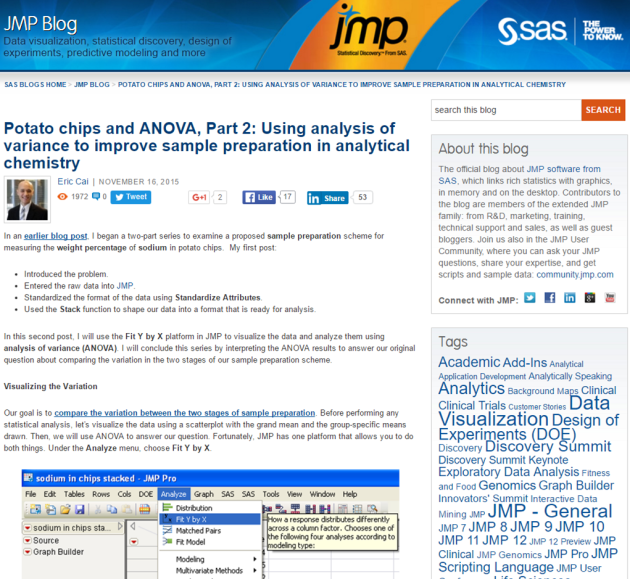
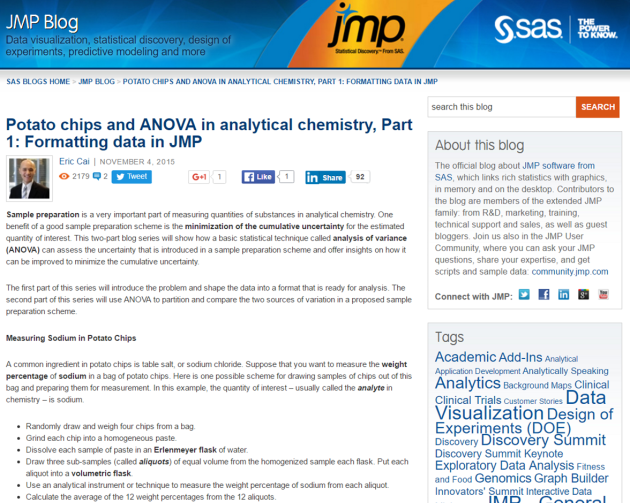
Yesterday, all 15 home teams in Major League Baseball won on the same day – the first such occurrence in history. CTV News published an article written by Mike Fitzpatrick from The Associated Press that reported on this event. The article states, “Viewing every game as a 50-50 proposition independent of all others, STATS figured the odds of a home sweep on a night with a full major league schedule was 1 in 32,768.” (Emphases added)

Screenshot captured at 5:35 pm Vancouver time on Wednesday, August 12, 2015.
Out of curiosity, I wanted to reproduce this result. This event is an intersection of 15 independent Bernoulli random variables, all with the probability of the home team winning being 0.5.
![P[(\text{Winner}_1 = \text{Home Team}_1) \cap (\text{Winner}_2 = \text{Home Team}_2) \cap \ldots \cap (\text{Winner}_{15}= \text{Home Team}_{15})]](https://s0.wp.com/latex.php?latex=P%5B%28%5Ctext%7BWinner%7D_1+%3D+%5Ctext%7BHome+Team%7D_1%29+%5Ccap+%28%5Ctext%7BWinner%7D_2+%3D+%5Ctext%7BHome+Team%7D_2%29+%5Ccap+%5Cldots+%5Ccap+%28%5Ctext%7BWinner%7D_%7B15%7D%3D+%5Ctext%7BHome+Team%7D_%7B15%7D%29%5D&bg=f0f0f0&fg=555555&s=0&c=20201002)
Since all 15 games are assumed to be mutually independent, the probability of all 15 home teams winning is just
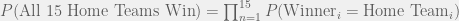
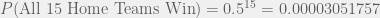
Now, let’s connect this probability to odds.
It is important to note that
- odds is only applicable to Bernoulli random variables (i.e. binary events)
- odds is the ratio of the probability of success to the probability of failure
For our example,
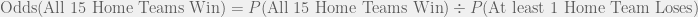

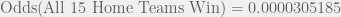
The above article states that the odds is 1 in 32,768. The fraction 1/32768 is equal to 0.00003051757, which is NOT the odds as I just calculated. Instead, 0.00003051757 is the probability of all 15 home teams winning. Thus, the article incorrectly states 0.00003051757 as the odds rather than the probability.
This is an example of a common confusion between probability and odds that the media and the general public often make. Probability and odds are two different concepts and are calculated differently, and my calculations above illustrate their differences. Thus, exercise caution when reading statements about probability and odds, and make sure that the communicator of such statements knows exactly how they are calculated and which one is more applicable.

Recent Comments